rpact: An R Package For Adaptive Clinical Trials
ROeS 2025, Graz
RPACT GbR
September 16, 2025
Overview 📦
- Comprehensive validated R package implementing methodology described in Wassmer and Brannath (2016)
- Enables the design of traditional and confirmatory adaptive group sequential designs
- Provides interim data analysis and simulation including early efficacy stopping and futility analyses
- Enables sample-size reassessment with different strategies
- Enables treatment arm selection in multi-stage multi-arm (MAMS) designs
- Enables subset selection in population enrichment designs
- Provides a comprehensive and reliable sample size calculator

Developed by RPACT 🏢
- RPACT company founded in 2017 by Gernot Wassmer and Friedrich Pahlke
- Idea: open source development with help of “crowd funding”
- Currently supported by 21 companies
- \(>\) 80 presentations and training courses since 2018, e.g., FDA in March 2022
- 29 vignettes based on Quarto and published on rpact.org/vignettes
- 28 releases on CRAN since 2018



RCONIS 🚀
- Grow RPACT company to offer a wider range of services
- Statistical consulting and engineering services: Research Consulting and Innovative Solutions
- Joint venture between RPACT GbR (rpact.com) and inferential.biostatistics GmbH (inferential.bio) founded by Daniel Sabanés Bové and Carrie Li
- Website: rconis.com

The R Package rpact – Functional Range

Trial Designs 🔬
- Fixed sample designs:
- continuous, binary, count, survival outcomes
- Group sequential designs:
- efficacy interim analyses, futility stopping, alpha-spending functions
- Adaptive designs:
- Inverse normal and Fisher’s combination test, conditional error rate principle
- Provides adjusted confidence intervals and bias corrected estimates
- Multi-arm multi-stage (MAMS) and enrichment designs, sample size reassessment
Sample Size and Power Calculation 💻
Sample size and power can be calulcated for testing:
- means (continuous endpoint)
- proportions (binary endpoint)
- hazards (survival endpoint)
- Note: flexible recruitment and survival time options
- rates (count endpoint)
Example: Sample Size Calculation 🧮
Sample size calculation for a continuous endpoint
Sequential analysis with a maximum of 3 looks (group sequential design), one-sided overall significance level 2.5%, power 80%. The results were calculated for a two-sample t-test, H0: mu(1) - mu(2) = 0, H1: effect = 2, standard deviation = 5.
| Stage | 1 | 2 | 3 |
|---|---|---|---|
| Planned information rate | 33.3% | 66.7% | 100% |
| Cumulative alpha spent | 0.0001 | 0.0060 | 0.0250 |
| Stage levels (one-sided) | 0.0001 | 0.0060 | 0.0231 |
| Efficacy boundary (z-value scale) | 3.710 | 2.511 | 1.993 |
| Futility boundary (z-value scale) | 0 | 0 | |
| Efficacy boundary (t) | 4.690 | 2.152 | 1.384 |
| Futility boundary (t) | 0 | 0 | |
| Cumulative power | 0.0204 | 0.4371 | 0.8000 |
| Number of subjects | 69.9 | 139.9 | 209.8 |
| Expected number of subjects under H1 | 170.9 | ||
| Overall exit probability (under H0) | 0.5001 | 0.1309 | |
| Overall exit probability (under H1) | 0.0684 | 0.4202 | |
| Exit probability for efficacy (under H0) | 0.0001 | 0.0059 | |
| Exit probability for efficacy (under H1) | 0.0204 | 0.4167 | |
| Exit probability for futility (under H0) | 0.5000 | 0.1250 | |
| Exit probability for futility (under H1) | 0.0480 | 0.0035 |
Legend:
- (t): treatment effect scale
Adaptive Analysis 📈
Perform interim and final analyses during the trial using group sequential method or p-value combination test (inverse normal or Fisher)
Calculate adjusted point estimates and confidence intervals (cf., Robertson et al. (2023), Robertson et al. (2025))
Perform sample size reassessment based on the observed data, based on calculation of conditional power
Some highlights:
- Automatic boundary recalculations during the trial for analysis with alpha spending approach, including under- and over-running
- Adaptive analysis tools for multi-arm trials and enrichment designs
Simulation Tool 🧪
Obtain operating characteristics of different designs:
- Assessment of adaptive sample size recalculation strategies
- Assessment of treatment selection strategies in multi-arm trials
- Assessment of population selection strategies in enrichment designs
Recent Updates

1. Count Data
getSampleSizeCounts()andgetPowerCounts()- Perform sample size calculations and the assessment of test characteristics for clinical trials with negative binomial distributed count data.
- Fixed sample size and group sequential designs (Mütze et al. (2019))
- Perform blinded sample size reassessments according to Friede and Schmidli (2010)
- Output boundary values also on the treatment effect scale
- New function
getSimulationCounts()- Perform power simulations for clinical trials with negative binomial distributed count data
- Simulated power, stopping probabilities, conditional power, and expected sample size for testing mean rates for negative binomial distributed event numbers in the two treatment groups testing situation
Count Data: Options
- Either both event rates \(\lambda_1\) and \(\lambda_2\), or \(\lambda_2\) and rate ratio \(\theta\), or \(\lambda\) and \(\theta\) (blinded sample size reassessment!)
- \(\lambda_1\) and \(\theta\) can be vectors
- Different ways of calculation:
- fixed exposure time,
- accrual and study time,
- or accrual and fixed number of patients
- Staggered patient entry
- Group sequential or fixed sample size setting
Count Data: Simple Example
Count Data: Simple Example
Sample size calculation for a count data endpoint
Fixed sample analysis, two-sided significance level 5%, power 90%. The results were calculated for a two-sample Wald-test for count data, H0: lambda(1) / lambda(2) = 1, H1: effect = 0.75, lambda(2) = 0.4, overdispersion = 0.5, fixed exposure time = 1.
| Stage | Fixed |
|---|---|
| Stage level (two-sided) | 0.0500 |
| Efficacy boundary (z-value scale) | 1.960 |
| Lower efficacy boundary (t) | 0.844 |
| Upper efficacy boundary (t) | 1.171 |
| Lambda(1) | 0.300 |
| Number of subjects | 1736.0 |
| Maximum information | 127.0 |
Legend:
- (t): treatment effect scale
Count Data: Power Calculation
Count Data: Power Simulation
$overallReject
[1] 0.835 0.0281.65 sec elapsed2. Delayed Response
- The delayed response group sequential methodology from Hampson and Jennison (2013) defines a proper consideration of the “pipeline” patients who have been treated at interim but are yet to respond
getDesignGroupSequential(),getDesignCharacteristics(), and the correspondinggetSampleSizexxx()andgetPowerxxx()functions characterize a delayed response group sequential test given certain input parameters in terms of power, maximum sample size and expected sample size- Other approaches can conveniently be handled with the newly published function
getGroupSequentialProbabilities() - Details and examples provided in Vignette Delayed Response Design with rpact.
Methodology of Delayed Response Designs
Given boundary sets \(\{u^0_1,\dots,u^0_{K-1}\}\), \(\{u_1,\dots,u_K\}\) and \(\{c_1,\dots,c_K\}\), a \(K\)-stage delayed response group sequential design has the following structure:


According to Hampson and Jennison (2013), the boundaries \(\{c_1, \dots, c_K\}\) with \(c_K = u_K\) are chosen such that “reversal probabilities” are balanced, to ensure type I error control.
More precisely, \(c_1,\ldots,c_{K - 1}\) are chosen as the (unique) solution of: \[\begin{align*} \begin{split} &P_{H_0}(Z_1 \in (u^0_1, u_1), \dots, Z_{k-1} \in (u^0_{k-1}, u_{k-1}), Z_k \geq u_k, \tilde{Z}_k \leq c_k) \\ &= P_{H_0}(Z_1 \in (u^0_1, u_1), \dots, Z_{k-1} \in (u^0_{k-1}, u_{k-1}), Z_k \leq u^0_k, \tilde{Z}_k \geq c_k). \end{split} \end{align*}\]
Delayed Response: Example
Delayed Response: Example
Sequential analysis with a maximum of 3 looks (delayed response group sequential design)
Kim & DeMets alpha spending design with delayed response (gammaA = 2) and Kim & DeMets beta spending (gammaB = 2), one-sided overall significance level 2.5%, power 80%, undefined endpoint, inflation factor 1.0514, ASN H1 0.9269, ASN H01 0.9329, ASN H0 0.8165.
| Stage | 1 | 2 | 3 |
|---|---|---|---|
| Planned information rate | 30% | 70% | 100% |
| Delayed information | 16% | 20% | |
| Cumulative alpha spent | 0.0022 | 0.0122 | 0.0250 |
| Cumulative beta spent | 0.0180 | 0.0980 | 0.2000 |
| Stage levels (one-sided) | 0.0022 | 0.0109 | 0.0212 |
| Upper bounds of continuation | 2.841 | 2.295 | 2.030 |
| Lower bounds of continuation (binding) | -0.508 | 1.096 | |
| Decision critical values | 1.387 | 1.820 | 2.030 |
| Reversal probabilities | <0.0001 | 0.0018 | |
| Cumulative power | 0.1026 | 0.5563 | 0.8000 |
| Futility probabilities under H1 | 0.019 | 0.083 |
Delayed Response: Plot

3. Conditional Performance Score
Performance scores combine different performance criteria in one value (e.g. Liu, Zhu, and Cui (2008), Wu and Cui (2012))
Evaluation perspectives: global and conditional
- global: looking at the properties of the adaptive design before the study starts
- conditional: looking at the properties of the adaptive design when the interim result suggests a trial continuation
Conditional Performance Score (cont’d)
- Idea of the conditional performance score:
- Evaluation of second stage sample size update
- Conditional power and sample size are random variables and therefore best described by location and variation measures
- Score by Herrmann et al. (2020) consists of four components (location and variation of conditional power and sample size)
- Score and its components take values between 0 and 1 (the higher the values, the better the performance)
- Usage of
getPerformanceScore()of a simulation object.
Conditional Performance Score: Example
# Initialize group sequential design with O'Brien-Fleming-boundaries
design <- getDesignGroupSequential(
kMax = 2,
typeOfDesign = "OF",
futilityBounds = 0,
bindingFutility = TRUE
)
# Perform simulation
maxNumberOfSubjects <- 200
n1 <- maxNumberOfSubjects * design$informationRates[1]
alternative <- c(0.2, 0.3, 0.4, 0.5)
design |>
getSimulationMeans(
alternative = alternative,
plannedSubjects = c(n1, maxNumberOfSubjects),
minNumberOfSubjectsPerStage = c(NA, 1),
maxNumberOfSubjectsPerStage = c(NA, 2*maxNumberOfSubjects),
conditionalPower = 0.8,
maxNumberOfIterations = 1e05,
seed = 123
) |>
# Calculate performance score
getPerformanceScore()Conditional Performance Score: Example
Performance
- Location sample sizes: 0.3042, 0.9516, 0.7936, 0.7353
- Variation sample sizes: 0.2591, 0.2138, 0.2318, 0.3094
- Sub-score sample sizes: 0.2817, 0.5827, 0.5127, 0.5223
- Location conditional power: 0.7254, 0.8150, 0.8897, 0.9402
- Variation conditional power: 0.3691, 0.4391, 0.5460, 0.6560
- Sub-score conditional power: 0.5472, 0.6271, 0.7178, 0.7981
- Performance scores: 0.4144, 0.6049, 0.6153, 0.6602
RPACT Cloud
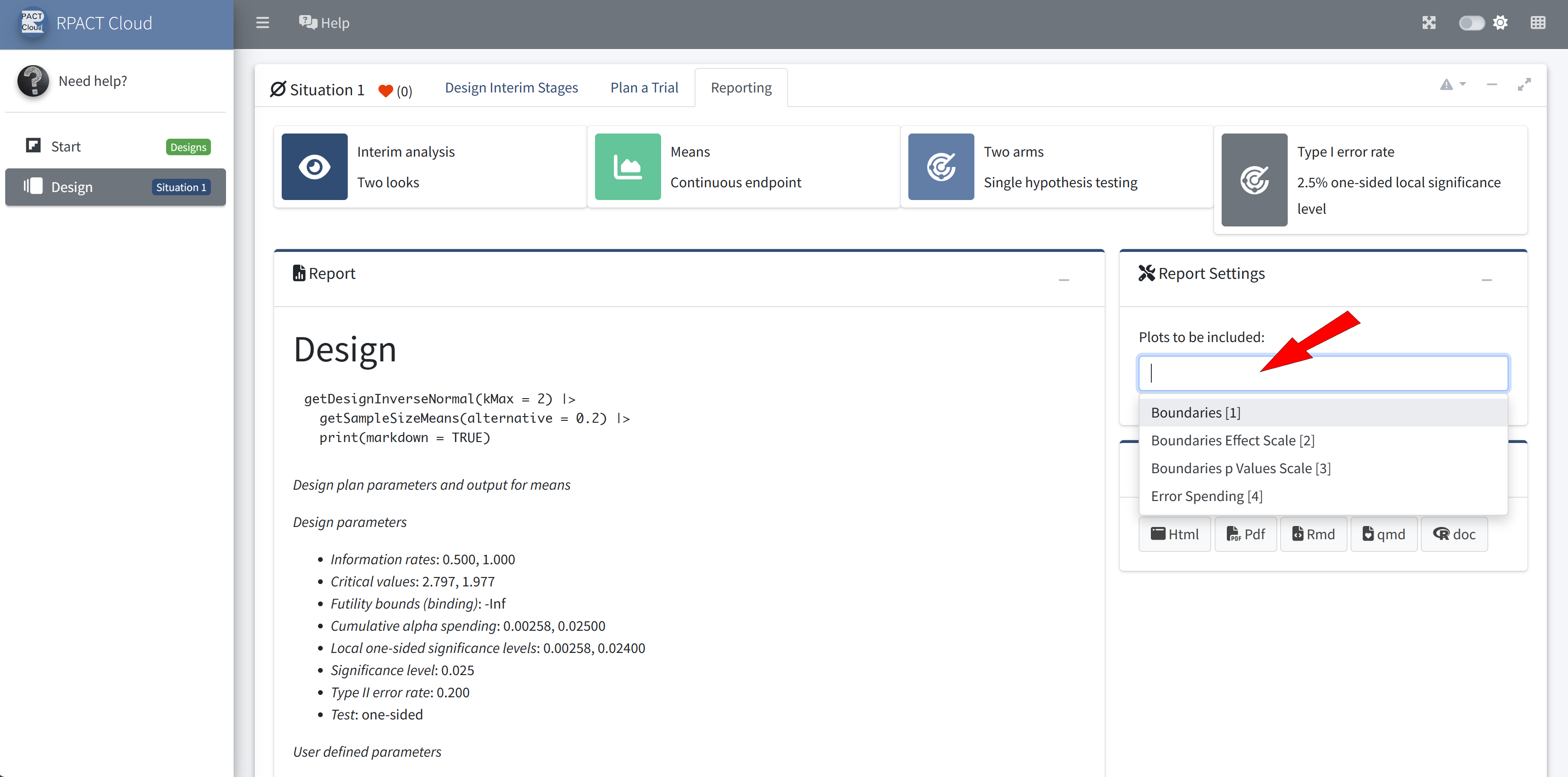
RPACT Cloud ☁️
Graphical user interface
Web based usage without local installation on nearly any device
Provides an easy entry to to learn and demonstrate the usage of
rpactStarting point for your R Markdown or Quarto reports
Online available at cloud.rpact.com
Start Page

Design

Reporting
What’s Next?

New Package Features
Just in the near future you will see in rpact:
- Patient level survival data simulations for multi-arm and enrichment designs
- Optimum conditional error functions according to Brannath et al. (2024) (package optconerrf published on CRAN Sep 09, 2025)
- Least square means interface for continuous endpoints
- Additional futility boundary scales: function
getFutilityBounds()- Available scales: zValue, pValue, conditionalPower, condPowerAtObserved, predictivePower, reverseCondPower, effectEstimate
Second Edition of the Book

- Provides up-to-date overview of group sequential and confirmatory adaptive designs in clinical trials
- Describes available software including R packages and has
rpactcode examples - Supplemented with a discussion of practical applications
The R Package rpact - Online Resources
Further information, installation, and usage:
- CRAN: cran.r-project.org/package=rpact
- GitHub: github.com/rpact-com/rpact
- GitHub Pages: rpact-com.github.io/rpact
- OpenSci R-universe: rpact-com.r-universe.dev/rpact
- METACRAN: r-pkg.org/pkg/rpact
- Codecov: app.codecov.io/gh/rpact-com/rpact
- RPACT manual and vignettes: rpact.org
- RPACT validation and training: rpact.com
All information and resources about RPACT on one dashboard page
RPACT Connect: connect.rpact.com
RPACT Connect
Thank you!
References
